Mechanistic analysis of antimicrobial peptide membrane interactions by molecular dynamics simulation
The world emergence of the bacterial superbugs has limited the effectiveness of the available antibiotics in the clinic. Because of the multifaceted antimicrobial mechanism of actions, cationic antimicrobial peptides (AMPs) may be considered as promising molecules to treat the infections associated with the resistant strains. AMPs are the molecules of the innate and adaptive immune systems of all organisms that save the host against wide varieties of microorganisms. They are 5-50 amino acids long and usually rich in the positively-charged arginine and lysine.

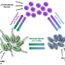
Fig. 1. Location of cationic AMPs in a) negatively b) neutral charged lipid membrane. The cationic amphiphilic peptides are fixed at the surface at the negatively charged membranes and the high amount of bound peptides is responsible for the disturbance of the bilayer architecture. An increase of the hydrophobic domain favors a deeper insertion into the hydrophobic interior of the membrane which consequently enhances the activity. Despite low binding to neutral membranes, AMPs may penetrate deeply into the hydrophobic region and disturb the arrangement of the acyl chains.
Despite the well-known membrane permeabilizing mode of action of the AMPs, the susceptibility of bacteria to AMPs depends upon both the peptide secondary structure and the physiochemical properties of the lipid membrane of bacterial cells (Fig. 1). Unlike the cholesterol rich electrically neutral lipid matrix of the membrane of normal eukaryotic cells; responsible for low peptide affinity and insertion, bacteria possess complex cellular envelope. In the case of gram-positive and negative bacteria, the membrane consists of a outer shells rich in the highly negatively charged lipopolysaccarides (LPS) and peptidoglycans (PEG), respectively, that represent together with the negatively charged lipids of the plasma membrane an ideal target to interact with AMPs.
Understanding the structure-activity relationship studies of AMPs along with the peptides’ mode of action forms the foundation for discovery of AMPs with unexploited mechanisms of action and improved therapeutic index against superbugs. On the level of the bacterial cell membrane; the electrostatic attraction favors binding between the cationic side chains of the AMPs and negative charges of the cytoplasmic lipid bilayers and the hydrophobic residues have a high propensity to insert into the membrane. Nevertheless, the effectiveness of the AMPs is proposed to be associated with factors that facilitate the transport of the peptide across the LPS and PEG. In this way, molecular dynamics (MD) simulation of the AMPs interactions with simple bacterial cytoplasmic lipid systems composed of appropriate molar ratio of the anionic; e.g., phosphatidylglycerol (PG), to the neutral; e.g., phosphatidylcholine (PC), phospholipids is beneficial to ensure the proper insight into AMPs’ mode of action.
In our recent paper; we explain a step-by-step protocol for the peptide structure modeling in water and bound to membrane using the Groningen machine for chemical simulations (GROMACS) package. We have taken advantages of small cationiccyclo(RRWFWR) peptide and simple 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) to discuss how to prepare a peptide-lipid complex for an MD run, extract the peptide structural stability and integrity parameters by clustering structures over the trajectory and measuring dihedral angles, and validate the lipid bilayer equilibration by analyzing for instance the area per lipid, bilayer leaflet thickness and lipid chains order parameters. We consider this paper important for the work of scientists involved in the basic as well as applied research on membrane active peptides.
Shima Arasteh, Mojtaba Bagheri
肽化学实验室,Institute of Biochemistry and Biophysics,
University of Tehran, Iran
Publication
Molecular Dynamics Simulation and Analysis of the Antimicrobial Peptide-Lipid Bilayer Interactions.
Arasteh S, Bagheri M
Methods Mol Biol. 2017












Leave a Reply
You must belogged into post a comment.