How do bacteria divide and multiply?
Bacteria interact with our bodies every day, resulting in both positive and negative outcomes. We rely on the billions of beneficial bacteria in our microbiome to support our digestion and immunity. At the same time, pathogenic bacteria can debilitate us when we’re exposed to just a few cells.
Understanding how bacteria divide from one cell into two daughter cells is crucial to designing ways to help promote or block multiplication of different bacterial species. Bacterial cell division, or cytokinesis, involves segregating replicated chromosomes so that each daughter cell gets one copy, and pinching the cell envelope closed between the two daughter cells.
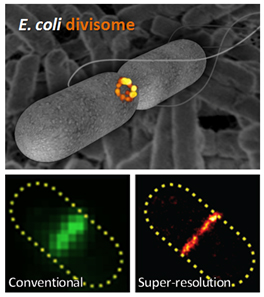
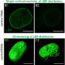
Fig. 1. (Top) Super-resolution fluorescence image of E. coli divisome protein, FtsZ, at midcell (orange) within a schematic cell outline (gray). Background: scanning electron micrograph of E. coli cells. (Bottom) FtsZ imaged in the same E. coli cell with conventional (left) or super-resolution (right) microscopy. Image credit: Carla Coltharp.
Many decades of study have provided a list of essential or ‘very important’ proteins (VIPs) required for cytokinesis, and revealed that these VIPs assemble into a ring-like ‘divisome’ at the middle of the cell (Fig. 1).
Our recent paper addressed one long-standing question about how the divisome functions: which VIP provides the driving force to propel cytokinesis along, thus dictating its speed? Knowing this power source would help us prioritize which proteins to target if we want to alter the speed of cytokinesis in certain bacteria.
To tackle this question, we first devised a very high-resolution microscopy method to get much sharper images of the divisome and bacterial cell boundaries, which appear fuzzy with conventional microscopes because bacteria are so small (Fig. 1). These super-resolution images allowed us to measure the speed of cytokinesis in bacterial cells much more precisely than we could before.
To tease out the relative importance of each VIP, we created different strains of bacteria with mutations that ‘broke’ each VIP, one at a time. We then applied our super-resolution methods to measure the rate of cytokinesis in each mutant strain, to see which mutations had the biggest effects on division speed.
We first tested mutations to a protein called FtsZ. FtsZ can form long polymers and has been proposed to be the power engine driving cytokinesis because it continuously releases chemical energy by breaking down a high energy molecule called GTP, much like the polymers of actin and myosin do during cell division in human cells. To our surprise, we found that mutations to FtsZ did not significantly change the rate of cytokinesis.
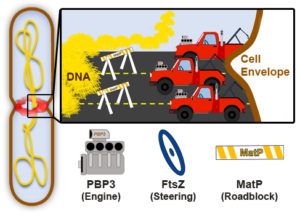
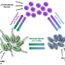
Fig. 2. Conceptual model of cytokinesis in bacteria. A dividing cell (left) contains separating DNA (yellow) and a midcell divisome (red), which inwardly constricts the cell envelope (brown). The speed and precision of cell envelope constriction is determined by the coordinated efforts of proteins such as PBP3, FtsZ, and MatP, depicted as participants in a driving scenario (right). Image credit: Ryan McQuillen and Carla Coltharp.
In contrast, we found that the rate of cytokinesis depended most on one VIP known as PBP3 (or penicillin binding protein 3). PBP3 is involved in building the cell wall that encases bacterial cells. When we impaired PBP3’s activity, cytokinesis slowed down significantly, suggesting that cell wall building may drive cytokinesis. This finding reveals a key difference between cell division in walled bacterial cells vs. wall-less animal cells.
Furthermore, when we severed a linkage between cell envelope-associated divisome proteins and chromosome-associated divisome proteins, we surprisingly discovered that cytokinesis proceeded faster. This protein linkage may thus function to check cytokinesis so that the envelope does not close in too fast before the two chromosome copies have finished separating.
Putting together our findings with those from many other groups, we arrive at a new picture of bacterial cytokinesis (Fig. 2). If we envision the cell envelope being towed by a car at midcell, PBP3 and the process of cell wall building would be the car’s engine, FtsZ would steer the car’s direction, and the chromosome linkage would impede forward progress when the envelope is in danger of running into unsegregated chromosomal DNA.
This new picture, together with the methods that we developed, opens up new avenues to explore the commonalities and specialized differences of cell division mechanisms across different bacterial species.
Carla Coltharp, Jie Xiao
Department of Biophysics and Biophysical Chemistry
Johns Hopkins School of Medicine
Baltimore, MD, USA
Publication
Defining the rate-limiting processes of bacterial cytokinesis.
Coltharp C, Buss J, Plumer TM, Xiao J
《美国国家科学院刊S a . 2016年2月23日









Leave a Reply
You must belogged into post a comment.