Heavy alcohol drinking and potassium channel genes
Alcohol use disorder is a chronic and relapsing brain disease that has damaging, sometimes deadly, consequences for the individual and costs society billions of dollars a year (approximately $223 billion). Despite alcohol use being one of the leading causes of preventable death in the United States, there are only a handful of FDA-approved drug treatments, all of which have had little success. In the age of personalized medicine and the absence of successful treatments, the focus of much research has shifted toward exploring the genetic basis of alcoholism in an effort to identify and validate new treatments. Recent evidence, from both preclinical and clinical studies, shows that a number of well-tolerated anti-seizure medications have great potential for treating AUD. We know that expression of the targets of some of these anticonvulsants, i.e., potassium channels, are altered in the brains of alcoholics and restoring normal function of these channels decreases consumption in rodent alcohol drinking models.

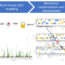
Fig. 2. Genetic variations in potassium channel gene expression in two integral brain regions in the reward system correlate with voluntary alcohol drinking in genetically diverse strains of mice. Mutations in potassium channel genes may alter expression and cell firing, ultimately impacting heavy alcohol drinking and alcohol dependence.
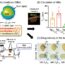
While this initial evidence is promising, the relationship between the genes that encode potassium channels and alcohol intake is poorly understood. Determining the connection between expression of these genes and drinking may provide valuable information for determining the efficacy of potential drug treatments. In order to experimentally assess these relationships, we analyzed genetic data in the GeneNetwork database (www.genenetwork.org) generated by the INIAstress Consortium, a group of NIH researchers dedicated to identifying the complex relationship between genes, alcohol, and stress. INIAstress investigators used the genetically diverse BXD recombinant inbred strains of mice in a model of alcohol dependence that enhances drinking in B6 mice (one of the parent strains of the BXDs). Once dependent on alcohol, INIAstress investigators performed genetic analyses of brain tissue samples from these mice. Using the genetic data, we correlated voluntary alcohol consumption with potassium channel gene expression levels in different parts of the brain’s reward system. We found that a number of potassium channel genes (e.g.,Kcnn,Kcnj, andKcnq) were altered by alcohol dependence and correlated with voluntary alcohol drinking in dependent and non-dependent BXD mice.
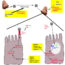
There is an FDA-approved anti-seizure medication (retigabine) that targets the voltage-gated potassium channels that are encoded byKcnqgenes. The availability of retigabine provided us with an opportunity to validate the role ofKcnqin a mouse model of heavy alcohol consumption. When we administered retigabine to B6 mice that had long access to alcohol, the high dose of retigabine only slightly reduced drinking. Even though the mice we used for this validation study were from the same strain, there was a lot of variability in their daily alcohol intake ranging from eight to 18 g/kg/day. Closer inspection of our results revealed that retigabine had no effect on alcohol consumption in low-drinking mice. However, both doses of retigabine tested significantly reduced drinking in the high-drinking mice, further validatingKcnqgenes as mediators of heavy drinking.
In summary, our bioinformatics analysis has shown that preclinical mouse models can be useful tools for identifying alcohol-sensitive potassium channel genes that may explain the genetic susceptibility of heavy or uncontrolled alcohol consumption. Our experimental validation ofKcnqas a novel pharmacogenetic target to regulate heavy alcohol drinking highlights the importance and usefulness of these tools. Because single nucleotide polymorphisms in humanKCNQ1andKCNQ5associate with early onset and symptoms associated with alcohol dependence, targetingKCNQhas substantial potential for advancing personalized medicine approaches toward treating alcohol use disorder.
Jennifer A. Rinker, Patrick J. Mulholland
Departments of Neuroscience and Psychiatry & Behavioral Sciences
Charleston Alcohol Research Center
Medical University of South Carolina, USA
Publication
Differential potassium channel gene regulation in BXD mice reveals novel targets for pharmacogenetic therapies to reduce heavy alcohol drinking.
Rinker JA, Fulmer DB, Trantham-Davidson H, Smith ML, Williams RW, Lopez MF, Randall PK, Chandler LJ, Miles MF, Becker HC, Mulholland PJ
Alcohol. 2017 Feb










Leave a Reply
You must belogged into post a comment.