Abiotic stress signaling in plants: functional genomic intervention
Basic plant science has never been more important than in the current scenario of climate change and increasing human population. To address the global challenges like increase in food production and raising stress tolerant crop varieties the basic understanding of plant biology is essential. To address these grand challenges, scientists are designing various tools and methodologies to study plant mechanisms and use that knowledge to devise field-based solutions.

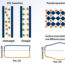
Fig. 1. The figure depicts various ‘Omics’ based approaches used to identify the function of a gene. Transcriptomics involves the analysis of mRNA abundance that corresponds to their expression in a sample. Phenomics involves analysis of the plants based on their appearance/morphology. Metabolomics is the study of nature of metabolites produced in a particular pathway. Proteomics is the study of the proteins and their activities. Interactomics involves the study of protein-protein interactions and their crosstalk to understand the correlation between them in a signaling pathway. All these approaches together are termed as ‘Functional Genomics’.
Plant biologists around the world are working to reveal the defense or tolerance mechanism indigenous to plants. These response mechanisms are highly complex and involve multiple signaling proteins. Besides performing independent functions, these signaling proteins also crosstalk to regulate number of physiological and developmental processes in plants. The function of these individual signaling proteins at the molecular level would enable plant biologist to engineer desired traits in agronomical and economically important plant species.
With the availability of whole genome sequences, next in line is the significant progress in methods related to gene expression, transcription and translation, also addressed informally as ‘Omics’ based approaches. These include transcriptomics, proteomics, metabolomics, interactomics and phenomics in several organisms (Fig. 1). All these approaches collectively are termed as ‘Functional Genomics’ that aims to explain the function of gene (s) and or gene family. Together these approaches have served immensely in the assessment and shaping of the molecular signaling network in the cells. The complete cellular map of signaling network is required to identify the proteins being functionally activated in response to a specific environmental stress signal.
Amongst the most common stresses faced by crop plants, are salinity, drought, cold and nutrient deficiency in the soil. Rice is the staple crop of major proportion of population worldwide. Rice is susceptible to salinity stress typically in coastal agricultural area. Water is an essential component for rice cultivation and factors such as climate change and low rainfall are depleting the water resources and leading to drought. Due to this, rice production is increasingly affected worldwide. The harmful effects of these stress conditions include low photosynthesis rate, poor growth of plants and reduce seed production.
The global research efforts by plant biologists have further gained momentum with the availability of whole genome sequences of many easy to study non-crop and crop plants. Not only this, a dramatic progress and use of techniques that can process and produce enormous amount of data have further improved our understanding of cellular response machinery. To know the target genes functioning under a particular stress conditions first of all ‘Genomics’ is used to locate the gene(s) and their sequence. Second, using the ‘Transcriptomics’ approach, expression of these genes under normal and stress conditions is assessed. Third, the highly responsive genes are then introduced into the plant and effect of transgene on the morphology of the plant (phenotype) is monitored using ‘Phenomics’. Next, to study the biochemical changes caused by transgene into plants ‘Metabolomics’ approach is employed. This approach help in identification and measuring the amount of small biomolecules generated as metabolic products. Further, to decipher the crosstalk or relation between genes within the same gene family or in particular signal transduction pathway ‘Interactomics’ approach is used. This approach aids in identification and confirmation of the protein-protein interactions within a cell. Lastly, to understand the biochemical structure, nature and activity of these proteins, ‘proteomics’ is used. This approach can further increase our understanding about the function of large set of proteins within a cell. Ultimately, by integrating the information from all the ‘Omic’ approaches, a complex problem can be tackled and pave the path for designing the future crops, which can adress the most important questions of food security and sustainability.
Manisha Sharma,Girdhar K. Pandey
Department of Plant Molecular Biology, University of Delhi South Campus,
Benito Juarez Road, Dhaula Kuan, New Delhi-110021, India
Publication
Editorial: Abiotic Stress Signaling in Plants: Functional Genomic Intervention.
Pandey GK, Pandey A, Prasad M, Böhmer M
Front Plant Sci. 2016 May 20











Leave a Reply
You must belogged into post a comment.