Two kinds of novel osmoregulation pathways in Escherichia coli
To cope with severe environmental changes, cell is equipped with various stress response pathways. Osmotic upshift (hyperosmotic shock) triggers water efflux from bacterial cell, and the resulting cell shrinkage is sensed by osmosensors embedded in the cytoplasmic membrane (Fig. 1.). This causes rapid influx of potassium ions through the transporters to counteract the osmotic pressure, and it also promotes the expression of the potassium transporter Kdp through the KdpD-KdpE two-component system. Concomitantly, glutamate gradually accumulates in the cell, and the accumulated potassium glutamate changes the general transcription governed by σ70to σS-specific transcription to promote uptake and synthesis of osmoprotectants (osmolytes) such as glycine betaine, proline and trehalose, which attenuate accumulation of potassium ions in the cell. The global change of gene expression for adaptation requires time: When high concentration of salt (around 0.9-1.0 M) is added to the culture medium, mid-log phase cells ofEscherichia coliimmediately stop growing and the growth resumes several hours later. Osmotic upshift stimulates another stress response pathway governed by σE, an alternative sigma factor that is involved in expression of periplasmic and outer membrane proteins. Thus, multiple pathways undergo regulation upon osmotic upshift, whereas involvement of the ribosome has not been focused.
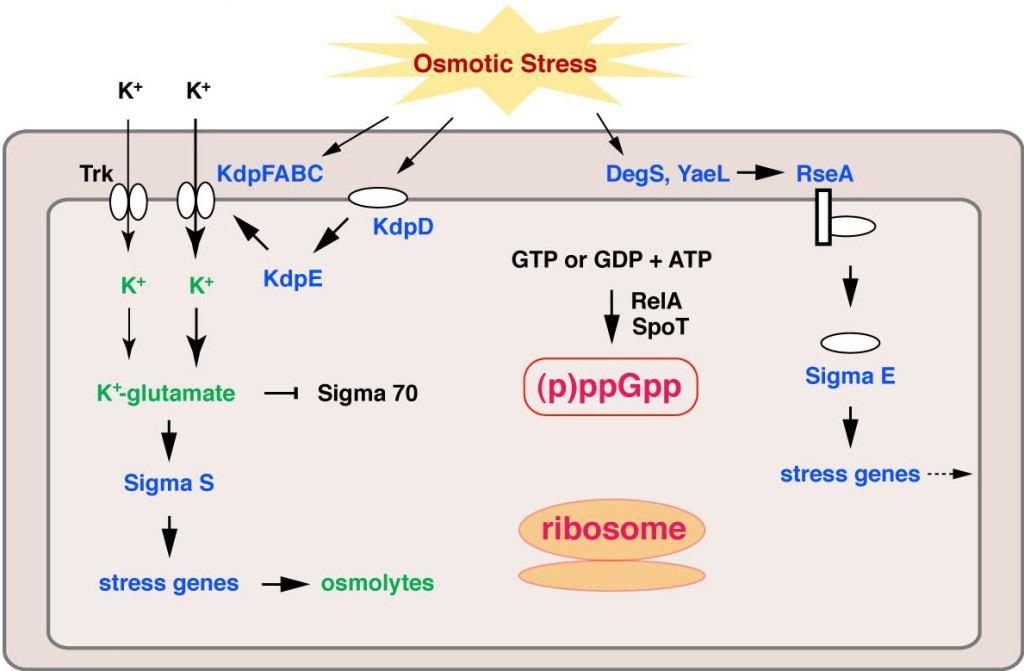
Fig. 1. Osmotic stress response pathways in E. coli. Factors or gene expressions up-regulated by osmotic shock are shown by blue, and metabolites and potassium ion accumulated in the cell after osmotic shock are shown by green.
We have previously shown thatE. colicells lacking a ribosome maturation factor resume growth earlier and grow faster after salt shock than do wild-type cells, although they grow slower under a normal condition. Earlier recovery of cell growth also occurs when some kinds of translation inhibitors are added in the cell culture. Taken together, impaired ribosome maturation or function providesE. colicells with resistance to salt stress. Although the underlying mechanism remains unknown, involvement of σEhas been suggested: σEis transiently induced after salt shock in cells defective in ribosome maturation or function earlier than in wild type cells.
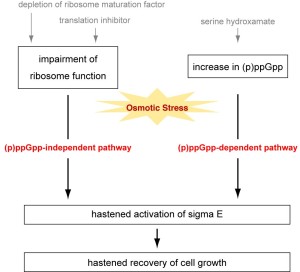
Fig. 2. Schematic representation of (p)ppGpp-dependent and -independent pathways for salt tolerance.
Here, we show another kind of osmoregulation pathway, which depends on ppGpp or pppGpp. (p)ppGpp is a bacterial second messenger that is produced from ATP and GTP or GDP by the stringent response factor RelA associated with the translating ribosome upon amino acid starvation. (p)ppGpp is produced by SpoT upon fatty acid starvation or phosphate limitation, while SpoT usually hydrolyzes (p)ppGpp into GTP or GDP and pyrophosphate. (p)ppGpp binds to RNA polymerase to change the binding specificity for s factors so that it serves as the global regulator of the gene expression. (p)ppGpp also binds to σE, IF2 and EF-G to modulate the gene expression pattern at the transcriptional and/or translational levels. We found that serine hydroxamate, which induces amino acid starvation leading to (p)ppGpp production, provides cells with salt resistance.
Depletion of ribosome maturation factor or addition of translation inhibitors is still effective in cells lacking (p)ppGpp, although serine hydroxamate is not. This indicates the presence of two kinds of novel salt response pathways (i.e., a (p)ppGpp-dependent pathway and a ribosome-related but (p)ppGpp-independent pathway) (Fig. 2). σS,另一个σ因子站转车nary phase or under various stressful conditions, is dispensable for either (p)ppGpp-dependent or -independent pathway. SHX causes hastened activation of σEafter salt shock, as observed in cells defective in ribosome maturation or function.
Hyouta Himeno, Simon GotoandTakefusa Tarusawa
Department of Biochemistry and Molecular Biology, Hirosaki University
Hirosaki, Japan
Publication
(p)ppGpp-dependent and -independent pathways for salt tolerance in Escherichia coli.
Tarusawa T, Ito S, Goto S, Ushida C, Muto A, Himeno H.
J. Biochem. 2016 Jan 27









Leave a Reply
You must belogged into post a comment.