How cells crawl… that’s right, crawl
Within our bodies, every cell must crawl – some only a few millionths of an inch, while others, like nerve fibers, must make their way from the spinal cord to the tips of our toes. The ability to crawl helps the cells of our skin to come together, keeping liquids within and pathogens without. When viewed under a microscope, a cell’s crawling motions – advancing testily, pausing occasionally, and withdrawing, when necessary – are what convince us it is alive.

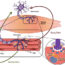
图1所示。行动的“积极”和“消极”actoclampin motors. Active end-tracking motors can harness the energy of on-filament ATP hydrolysis to generate high force on a biologically relevant time-scale (milliseconds), whereas passive tracking motors generate low forces because they lack any mechanochemical linkage to ATP hydrolysis. See text for other details.
This review describes how Richard Dickinson and I came to discover that cells are propelled by molecule-sized motors that form when a tracking protein binds to the tip of an actin filament, the latter a polymer formed from actin monomers. Like DNA, these filaments have a double-helical structure consisting of two strands that twist around each other; however, the strands of an actin filament are identical and run in the same direction. One end, called the (+)-end, is where actin monomers add. Importantly, the two strands are offset by half the length of a monomer, such that there is always a leading sub-filament and a lagging-subfilament. Each end-tracking motor is a multi-arm protein complex that employs one arm to recruit/deliver actin monomers to a growth-site located at the end of the lagging subfilament, while a second arm is bound at the end of the leading subfilament. Once a subunit adds, the lagging subfilament becomes the leading subfilament, and the arms trade functions. It is this alternating action that defines a true molecular motor, one that generates molecular-scale pushing forces, while remaining in full possession of the cargo it propels. The review also acknowledges an earlier notional model for actin polymerization motors suggested by Uno Lindberg and colleagues on the basis of electron microscopy images of near-membrane structures in highly motile cells.

Fig. 2. Cartoon showing the features of actoclampin motors, wherein an oligomeric end-tracking protein complex (shown in gray) maintains its purchase on the cargo-bound protein and has a force-generating link to the (+)-end of an elongating actin filament. Cargo includes: the cell’s leading edge (lamellipodium), a finger-like cell extension (filopodium), an invasive cell protuberance (invadipodium), a membrane vesicle (endosome or phagosome), or an intracellular pathogen (Listeria, Shigella, vaccinia, myxovirus, etc.). In the case of Listeria monocytogenes, 80-100 end-tracking motors form a propulsive ensemble.
Ensembles of filament end-tracking motors generate the forces for cell crawling, and the Actoclampin Model explains how these motors identify their cargo, hold fast to it, and generate the force needed to deliver its cargo where and when it is needed. It also shows how end-tracking proteins can generate greater force, when their motor actions are coupled to the free energy of ATP hydrolysis. In the “Lock, Load & Fire” scheme (Fig.1A), the motor cycles through several steps of an autocatalytic process.Lockingdescribes how one arm of a tracking protein is firmly attached to an ATP-containing subunit (red chevron) at the extreme end of an actin filament. In theLoadingstep, a polymerization cofactor, known as profilin (light blue circle), ushers an actin monomer (as Profilin·Actin·ATP) from a docking site on the end-tracking protein to an empty growth site (light gray outline) on the filament. Once accomplished, monomer loading triggersFiring, wherein ATP hydrolysis forms filament-bound Actin·ADP (dark blue), using the energy liberated to drive the release of the tightly bound arm, thereby setting up the system for another round of monomer addition. While a motor using theLL&Fmechanisms can generate upwards of 17-18 pN, other end-tracking motors operate passively (Fig 1B), lacking the means to harness the energy of ATP hydrolysis and generating only 2-3 pN through monomer addition alone.
Humans possess upwards of twenty-four different end-tracking proteins, each with similar features (Fig. 2). Mutations or other changes in end-tracking proteins give rise to renegade motors, unbridling cancer cells to migrate and allowing them to enter the bloodstream and disseminate in the process known as metastasis. Learning how to drug these motors promises new ways to defeat cancer.
Dan Purich
Professor of Biochemistry & Molecular Biology
Department of Biochemistry & Molecular Biology, University of Florida College of Medicine
Publication
Actoclampin (+)-End-tracking Motors: How the Pursuit of Profilin’s Role(s) in Actin-based Motility Twice Led to the Discovery of How Cells Crawl
Purich DL.
Biophys Chem. 2016 Feb










Leave a Reply
You must belogged into post a comment.